Institute of Physics, Academia Sinica
Polymer Physics and Complex Fluids Group
Research
Coarse-grained model for DNA dynamics on the micron / millisecond scale
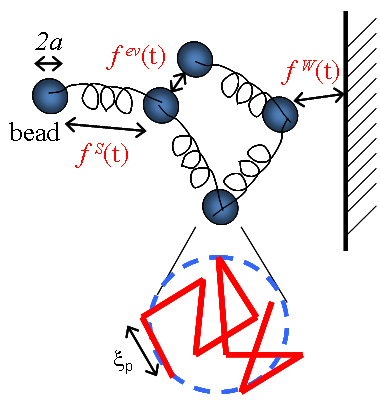
To capture the dynamics of DNA molecules on the micro length scale and second time scale, we use a coarse model that neglects the atomic structure of DNA, but captures its mechanical elasticity and its dynamics in fluid. [1] Using meso-scale simulations, a DNA molecule is represented by a bead-spring model with worm-like chain springs, where the spring force law comes from experimental measurements.
The hydrodynamic radius, excluded volume parameter, Kuhn length are matched to the measured relaxation time, chain diffusivity, and equilibrium stretch of lambda DNA. The hydrodynamic radius of each bead is 0.7 microns, and each bead is a coarse-grained representation of a random walk chain of ~ 20 Kuhn segments.
Comparisons between the model predictions with experiments show that the model captures DNA diffusivity in the bu lk fluid and in a confined environment. [2]
Here is a live demo
[1] R. M. Jendrejack, J.J. de Pablo, M.D. Graham (2002) Stocahstic simulation of DNA in flow: Dynamics and the effects of hydrod ynamic interactions, J. Chem. Phys. 116, 7752.
[2] Y.-L. Chen, M.D. Graham*, J.J. de Pablo*, G.C. Randall, M. Gupta, and P.S. Doyle* (2004) Conformation and Dynamics of Single DNA in Parallel-Plate Slit Microchannels, Phys. Rev. E, 70, 060901(R).