Computational study on how mechanical action of molecules influences fluctuating dynamics in the chromatin in a living cell
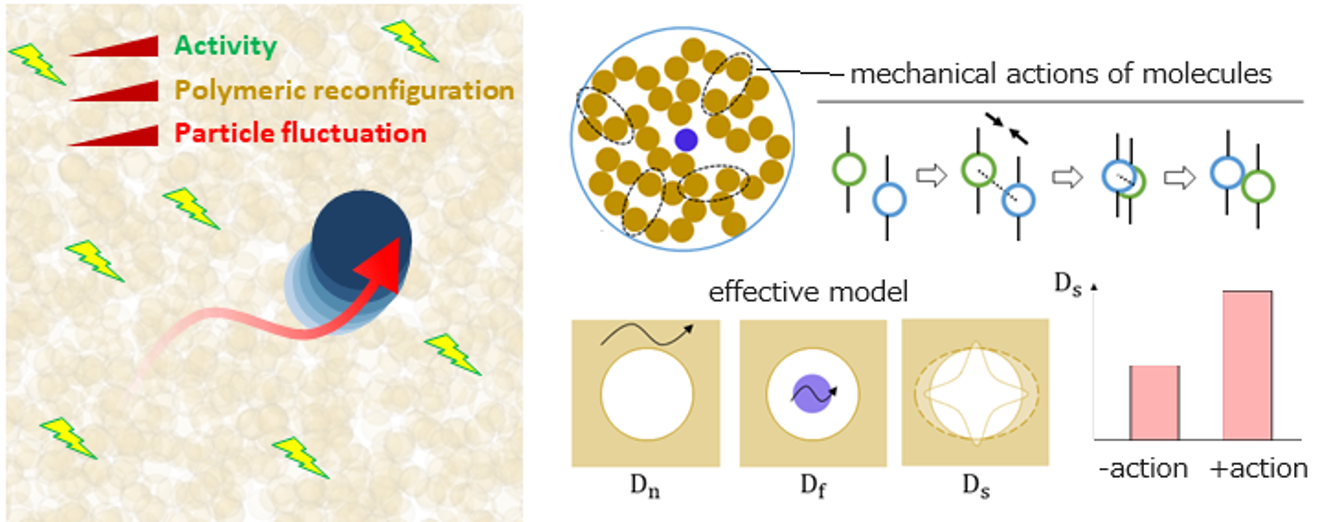
Genetic information of a eukaryotic cell is stored in its chromatin, a polymer-like entity comprising DNA and histone proteins, densely packed inside the nucleus. The nucleus also contains varieties of subnuclear compartments, whose spatiotemporal coordination relative to the chromatin is known to be an important factor determining the gene expressions. Meanwhile, in a living cell, various other subnuclear molecules like enzymes (such as polymerases and topoisomerases) should be acting to achieve cellular functions, and the mechanical perturbation by such acting molecules may affect fluctuating dynamics of subnuclear compartments in the chromatin. The international collaboration team led by Dr. Tetsuya Hiraiwa (IoPAS, and formerly Mechanobiology Institute in Singapore) and Dr. Jacques Prost (Curie Institute in Paris) numerically demonstrated that some classes of perturbations indeed enhance the fluctuation of the particles (mimicking the subnuclear compartments) within the complex polymeric medium (mimicking chromatin). They also clarified that this fluctuating dynamics can be effectively described by a combination of three dynamical modes, including a newly-discovered mode associated with the dynamic reconfigurations of polymeric meshes. This insight provides a fresh perspective on how mechanical perturbation of molecules may govern gene expressions by regulating the subnuclear-compartment dynamics. The complete work has been published recently on Physical Review Letters, Vol. 132, 058401 (2024).
Journal Links: https://journals.aps.org/prl/abstract/10.1103/PhysRevLett.132.058401